Super Resolution and Single Particle Fluorescence Microscopy
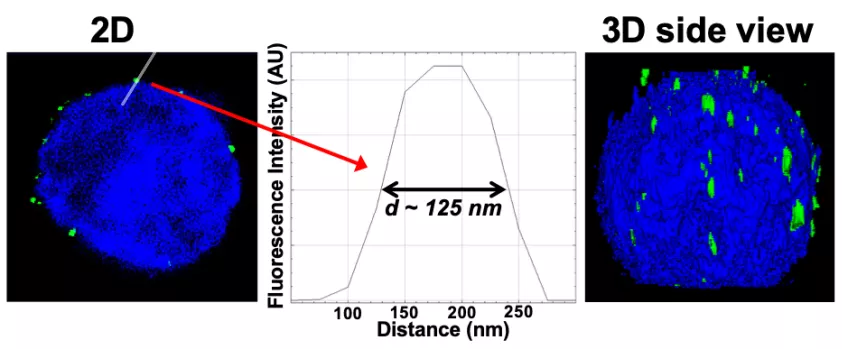
Super-Resolution Structured Illumination Microscopy (SR-SIM) provides resolutions down to 120 nm, allowing visualization of individual herpesvirus capsids (HSV-1) attached to the nucleus (HSV-1 C-capsid diameter is 125 nm).
Figure: Imaging of reconstituted capsid-nuclei system confirms specific capsid binding to the NPCs at the nuclear membrane. Representative super-resolution SIM image showing GFP-HSV-1 C-capsids (green) bound to isolated reconstituted rat liver nuclei (blue DAPI stain). A histogram of a capsid cross-section profile for a capsid GFP signal along the white line shows that individual C-capsids are resolved (HSV-1 C-capsid diameter ≈ 125 nm).
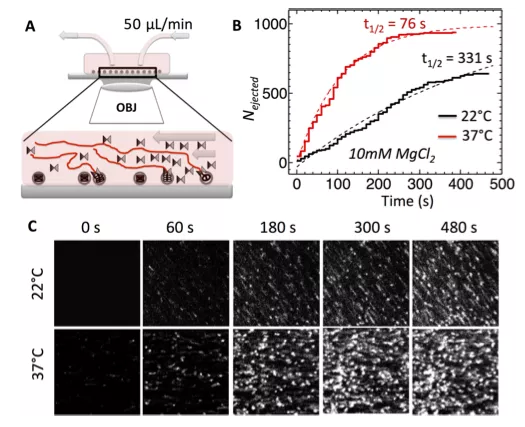
In parallel, using spinning disk confocal fluorescent microscopy (FM), single-molecule measurements, we investigate the average ensemble kinetics of the number of viruses which ejected their DNA versus time during an infection. This allows investigation of virus population dynamics.
Figure: Single-molecule fluorescence measurements of the ensemble kinetics for DNA ejection from WT phage λ over time in 10 mM MgCl2 Tris-buffer at 22 and 37°C. YOYO dye shows particles with ejected DNA stretched in the flow. The ejection is triggered by LamB receptor addition at time 0 and flown continuously with YOYO in the flow chamber.